Research
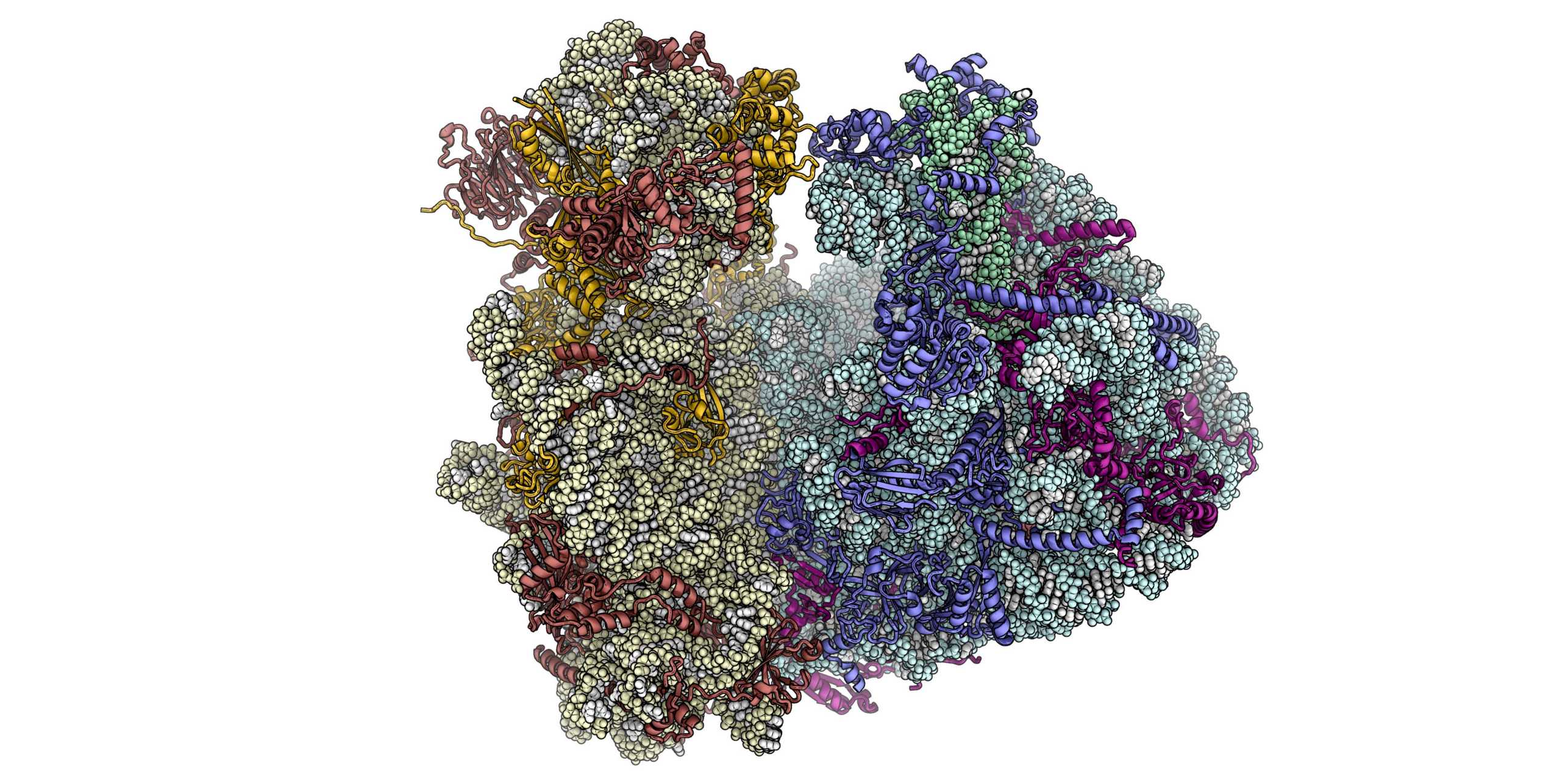
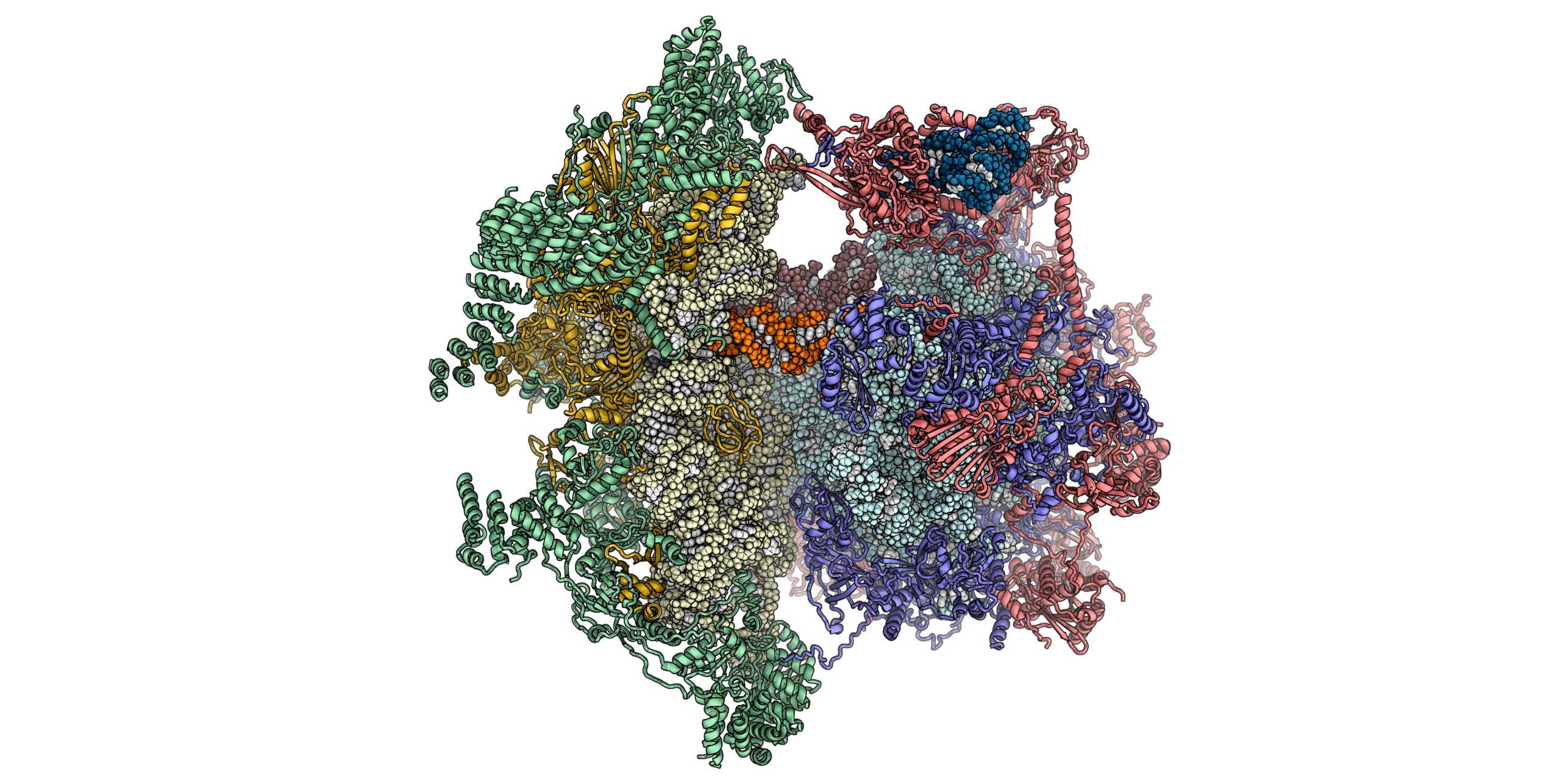
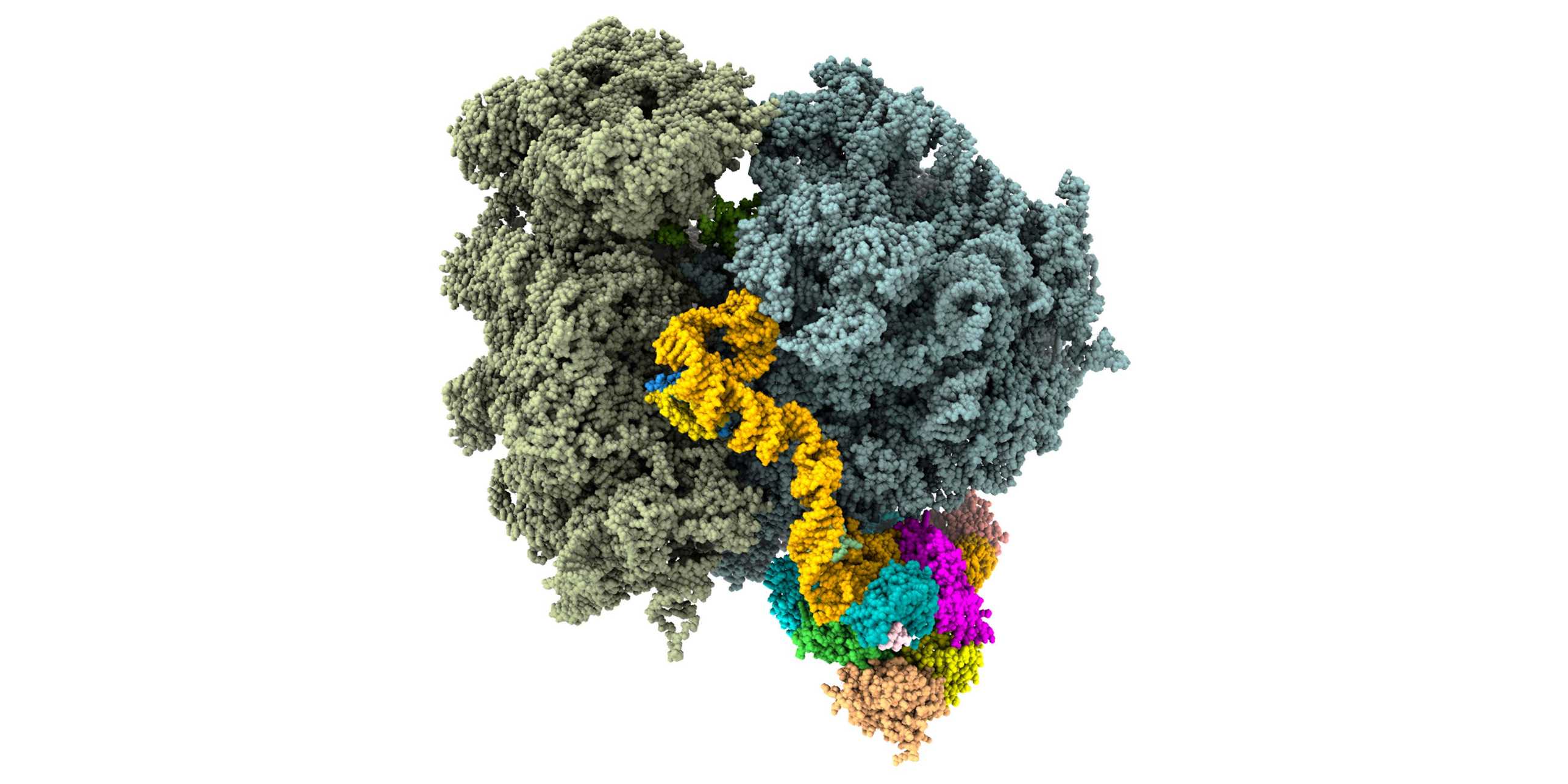
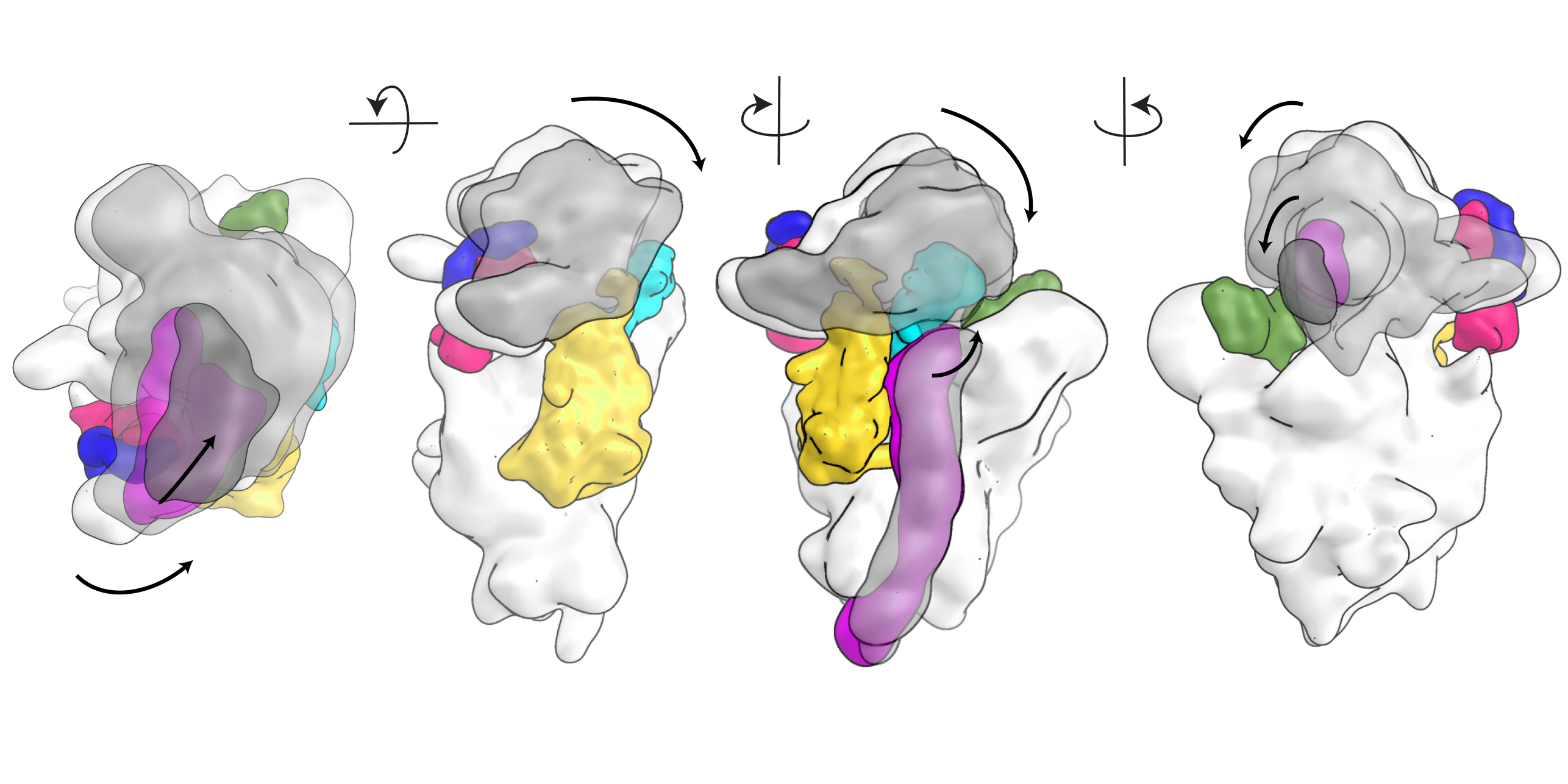
We are interested in understanding ribosomal structure and function in all kingdoms of life. Using a combination of biochemical, crystallographic and electron microscopic experiments we have obtained insights into co-translational nascent chain folding, processing and targeting to the membranes. Our recent focus is to understand eukaryotic cytoplasmic and mitochondrial ribosomes and their functional complexes involved in regulation and initiation of protein synthesis. Ultimately we intend to obtain a “molecular movie” of these processes at high resolution so that they can be understood at a detailed mechanistic level.
Eukaryotic Cytosolic Ribosomes
The eukaryotic protein synthesis machinery is much more complex than in bacteria. We have revealed the unique architectural features of eukaryotic ribosomes and are currently investigating the eukaryotic-specific mechanisms of translation initiation, ribosome assembly, and targeting of proteins to membranes.
Mitochondrial Protein Synthesis
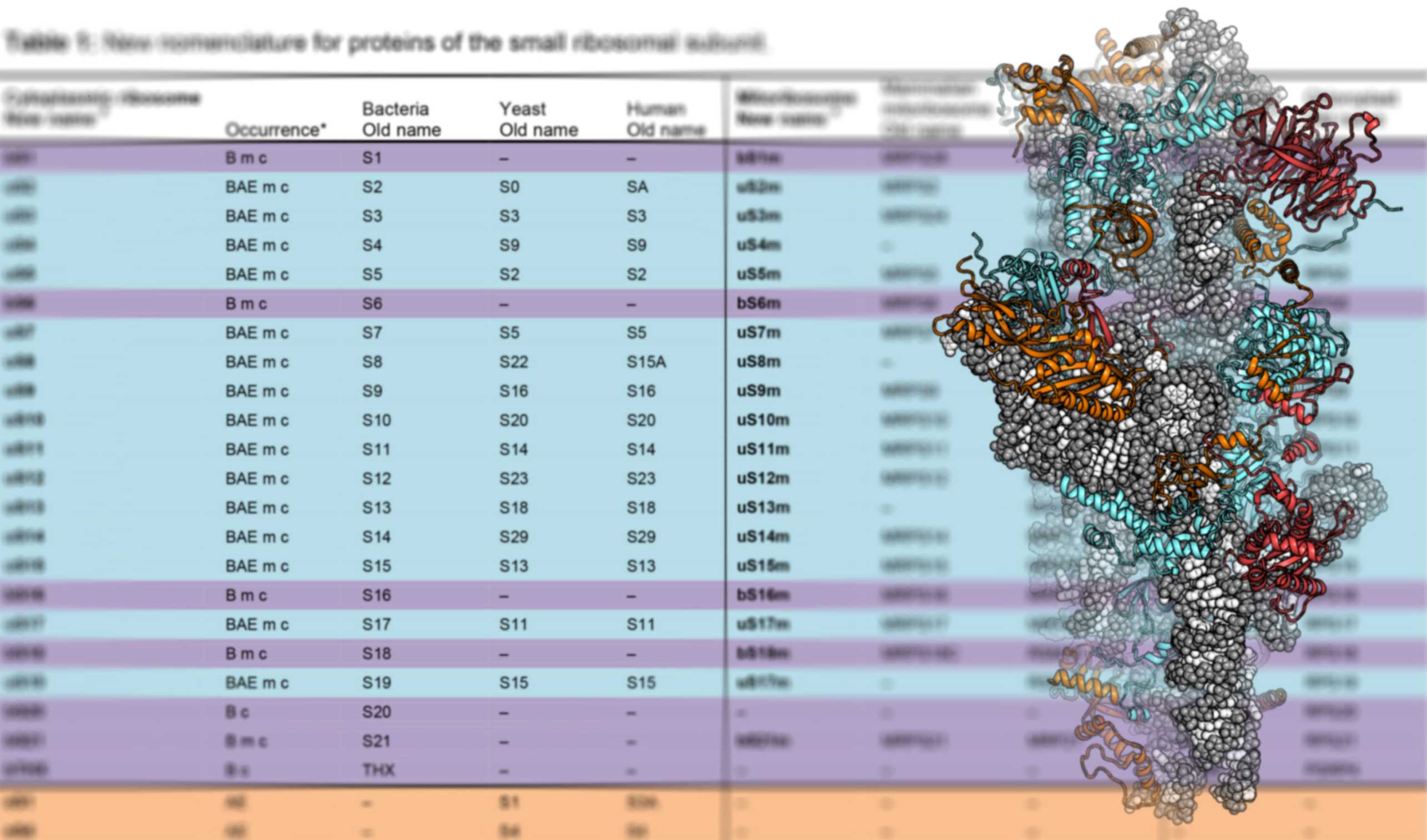
Mitochondria and chloroplast organelles in eukaryotic cells originated from free-living bacterial ancestors. Whereas ribosomes in chloroplasts are structurally still closely related to their bacterial ancestor, mitochondrial ribosomes diverged considerably during evolution, as visualized by our group for mammals and trypanosomes.