Co-translational Protein Targeting, Processing, and Folding
Cotranslational protein biogenesis and targeting in eukaryotes
During protein synthesis ribosomes are responsible for template-based polymerization of amino acids according to the information encoded in mRNA. Newly synthesized proteins diffuse from the ribosomal active site through a nascent polypeptide exit tunnel. The exit of the ribosomal tunnel can be considered a platform where different factors bind to control protein folding, processing and targeting to membranes. The participation of these factors is spatially and temporally coordinated to ensure faithful protein production and accurate cellular localization. Although these processes are of key importance for all cellular functions, given their complexity, the roles of many participating factors and the nature of their interplay at the exit of the ribosomal tunnel remain enigmatic.
We are investigating the molecular mechanisms of various cellular factors that act on the ribosome to facilitate protein production, with a particular focus on the mammalian system. Using a combination of biochemical and structural experimental approaches we study i) the temporal control of binding, i.e. at which point during protein synthesis different nascent chain-associating factors bind and in which order, ii) the spatial control of binding, i.e. where on the ribosome the factors bind and the implications of the overlap of binding sites for their temporal exchange, iii) how the factors cooperate, i.e. which ones bind simultaneously and how they interact with each other with implications for understanding the handover of nascent chains during protein production.
NAC recruits METAP and controls methionine excision
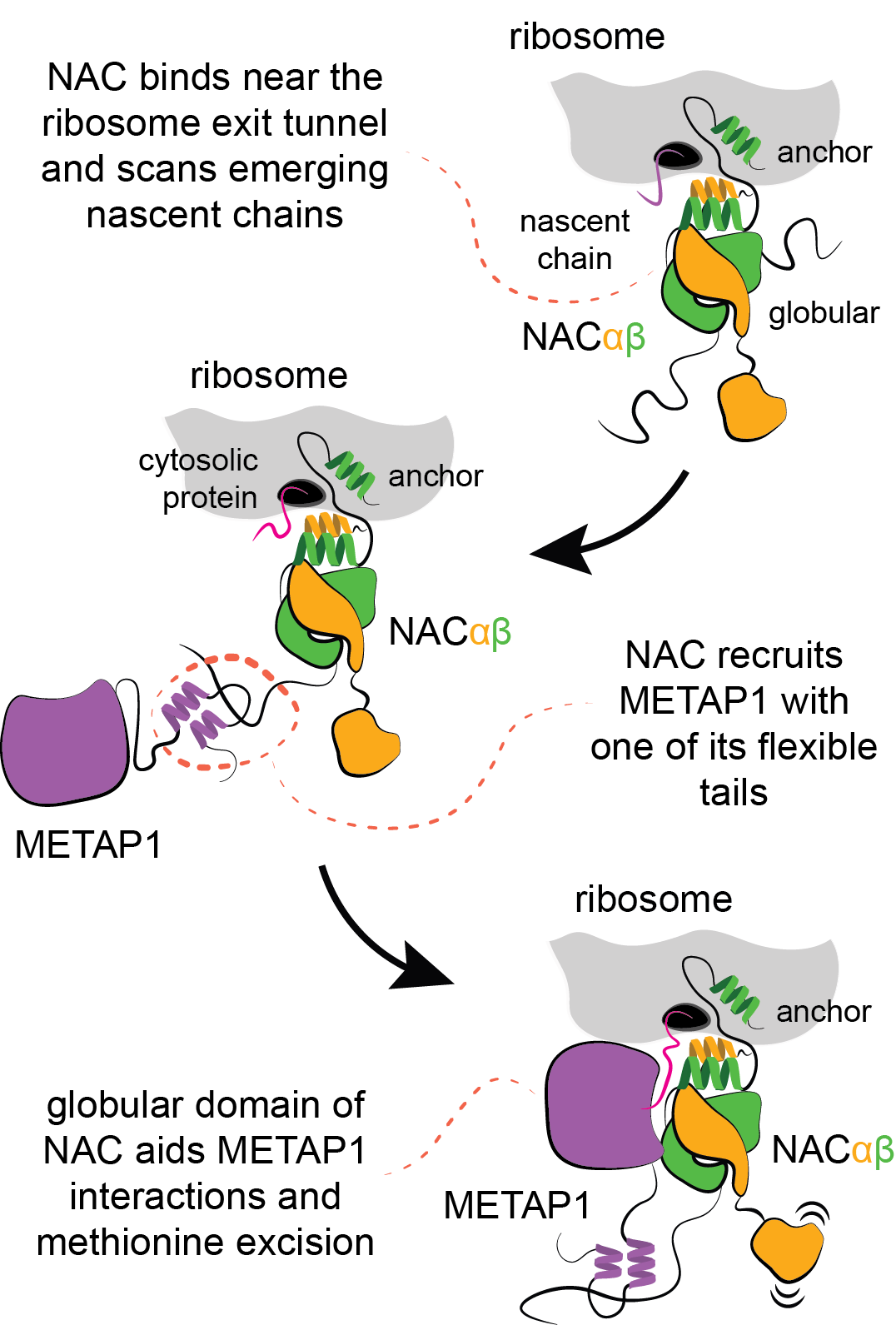
Our recent discoveries reveal that in eukaryotes, human cells for example, the nascent polypeptide–associated complex (NAC) controls all aspects of co-translational nascent chain biogenesis. NAC is a dimeric protein with a globular core and flexibly tethered tails. One of the tails serves as an anchor to position the globular domain above the exit of the ribosomal tunnel, whereas the other three tails are available for a multitude of interactions.
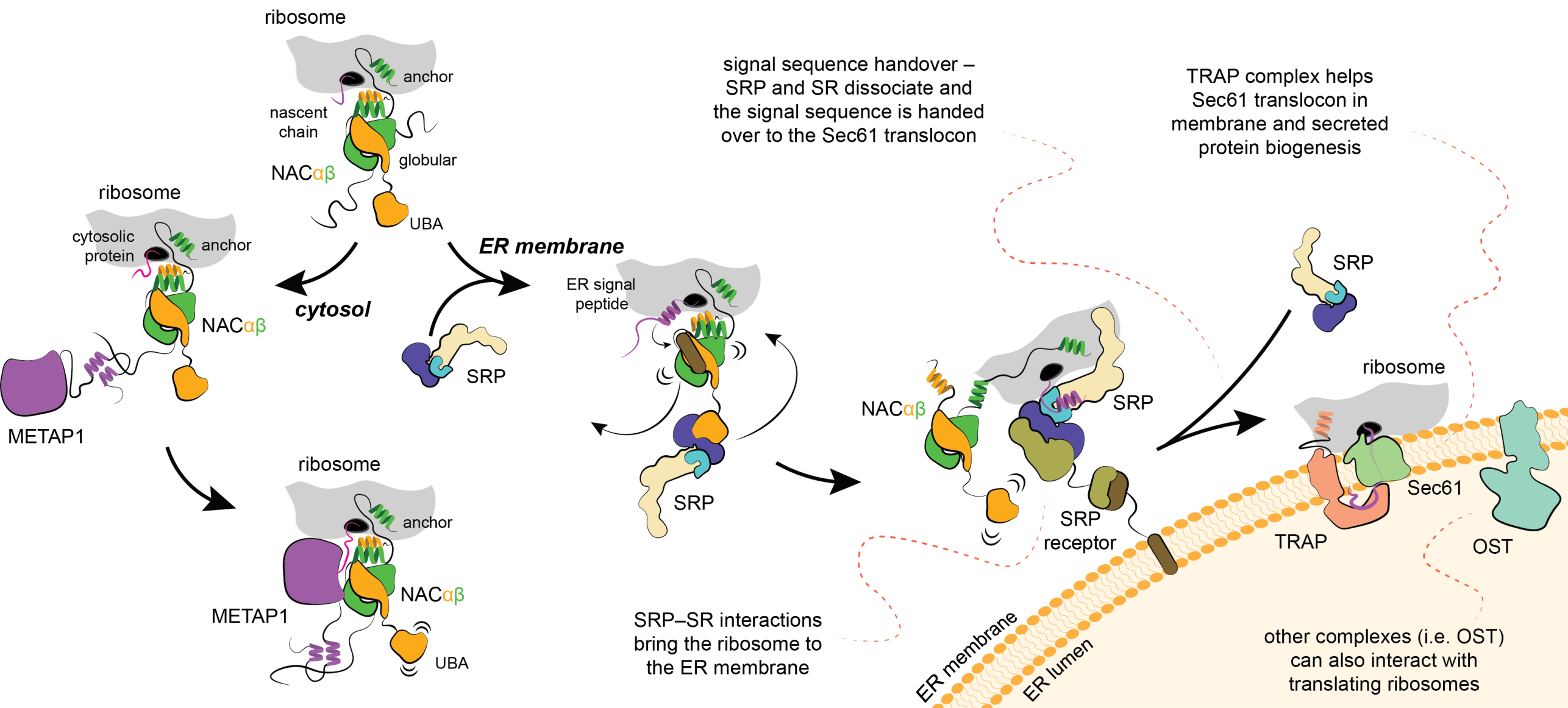
Methionine, the first amino acid incorporated in all newly synthesized proteins, is excised with a methionine aminopeptidase enzyme (METAP). In collaboration with the group of Elke Deuerling at Konstanz (LINK) we discovered that NAC recruits METAP via one of its flexible tails facilitating productive docking of METAP at the ribosome for nascent chain processing (external page Gamedinger et al 2023). Importantly, the functional association of METAP with NAC is dependent on interactions with the globular domain which stability is dependent on the type of nascent chain being translated. When cytosolic proteins are synthesized, the globular domain of NAC facilitates METAP interactions and methionine excision.
ER targeting is mediated by SRP and its interplay with NAC
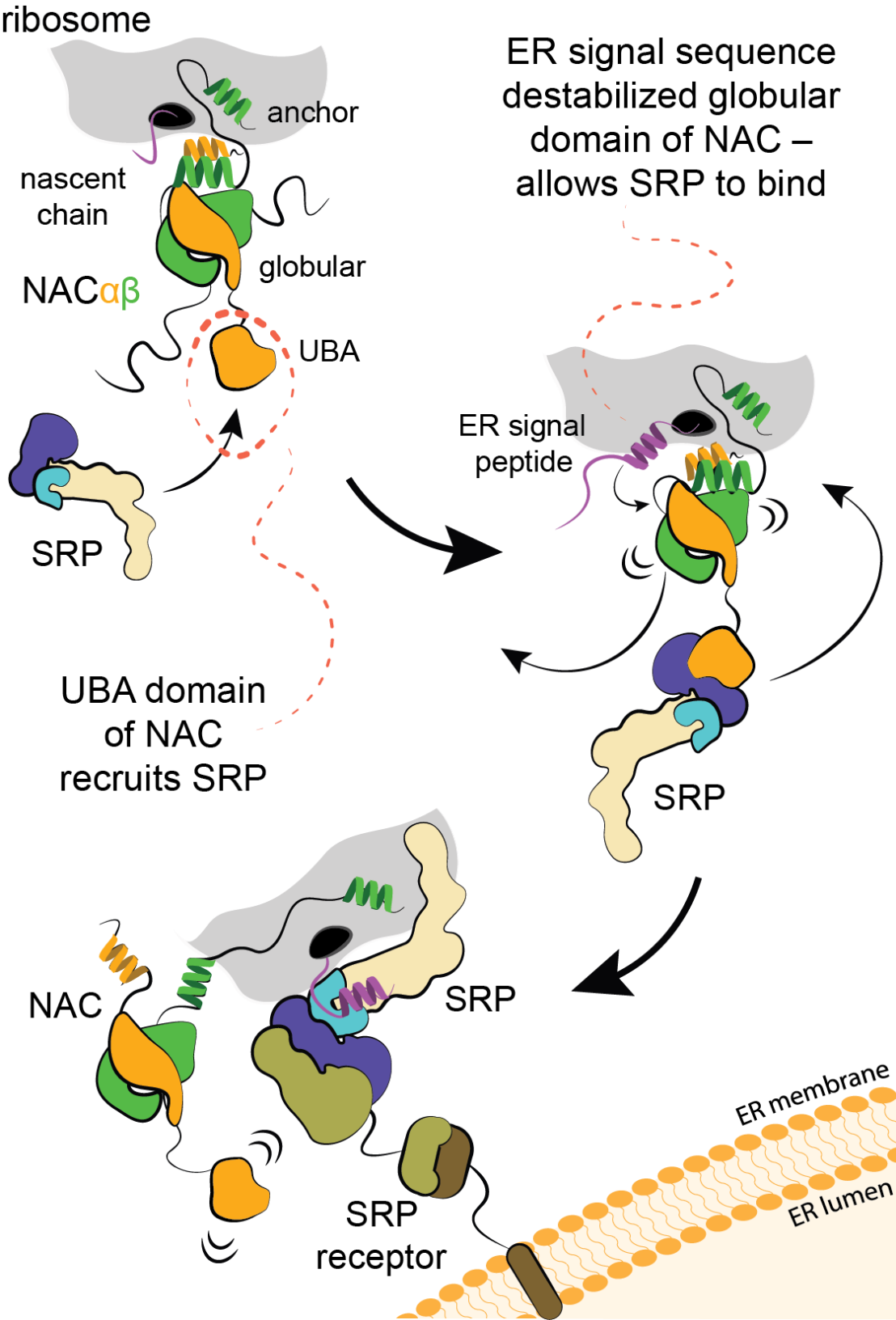
In collaboration with external page Shu-ou Shan’s group at Caltech (LINK) and the group of external page Elke Deuerling at Konstanz, we discovered that a different flexible tail of NAC, with a small UBA domain at its end, recruits signal recognition particle (SRP), which is responsible for cotranslational targeting of proteins to the ER. In contrast to the situation with METAP, the globular domain of NAC prevents SRP binding. However, when hydrophobic signal sequence emerges from the ribosomal tunnel, the globular domain of NAC becomes destabilized, allowing binding of SRP (external page Jomaa et al., 2022). Therefore, NAC critically contributes to the fidelity of targeting to the ER. At the ER, SRP engaged ribosome will bind to the flexibly disposed GTP-binding domain of the SRP receptor (SR). These interactions occur far away from the surface of ER, however, once SRP engages SR, the interacting domains are relocated to a distal region of the SRP (external page Kobayashi et al., 2018) and the long linker of SR retracts to bring the ribosome to ER (external page Jomaa et al., 2021; external page Lee et al., 2021). These conformational changes position the ribosome above the ER in a conformation that allows signal sequence handover to the protein conducting pore in the membrane, so called Sec61 translocon.
TRAP complex aids translocon in membrane and secreted protein biogenesis
Membrane proteins delivered cotranslationally to the ER will be inserted into the membrane through the lateral gate of Sec61. In case the protein needs to be translocated across the membrane, which is the case for many hormones, including insulin, the process will be facilitated by the TRanslocon Associated Protein (TRAP) complex. In collaboration with the group of external page Elke Deuerling we discovered that TRAP uses a flexibly disposed tail that binds to the ribosome as it approaches the ER membrane. Additional interactions help orient TRAP to position a cradle-like lumenal domain below the pore of the translocon where it can act as a chaperone to facilitate folding and biogenesis of translocated proteins (external page Jaskolowski et al. 2023).
Harnessing the power of 3D animation to visualize cotranslational events
The video below is an artistic interpretation of co-translational methionine cleavage by MetAP1 (see above) and SRP-directed membrane targeting and handover. It is based on a wealth of research from ours and other groups that contributed to a complete picture of this process. A lot of intermediate states of these dynamic processes have been solved as static structures, which the animation was based upon. However, in order to provide the necessary clarity, many processes have been simplified.