Mitochondrial Protein Synthesis
Mitochondrial Protein Synthesis
Mitochondria are organelles that are responsible for energy conversion in eukaryotic cells. The origin of mitochondria traces back to a free-living bacterium and to this day, mitochondria retain key features of their bacterial ancestors. This includes a separate mitochondrial genome and the complete molecular machinery to express the therein encoded genes. A central part of this machinery is the mitochondrial ribosome (mitoribosome) that is responsible for the synthesis of the mitochondrially encoded proteins.
Due to their evolutionary origins, mitoribosomes are more closely related to bacterial ribosomes than to eukaryotic cytosolic ribosomes. However, they have undergone extensive structural and compositional remodeling throughout evolution, which is reflected in striking variations in the rRNA content and the acquisition of a large number of mitochondria-specific ribosomal proteins. Mitoribosomes are highly specialized, synthesizing mostly membrane proteins that form parts of the protein complexes of the mitochondrial respiratory chain. Consequently, misfunctioning of mitochondrial translation is involved in a range of human pathologies such as cardiomyopathies, developmental abnormalities, cancer and hearing loss.
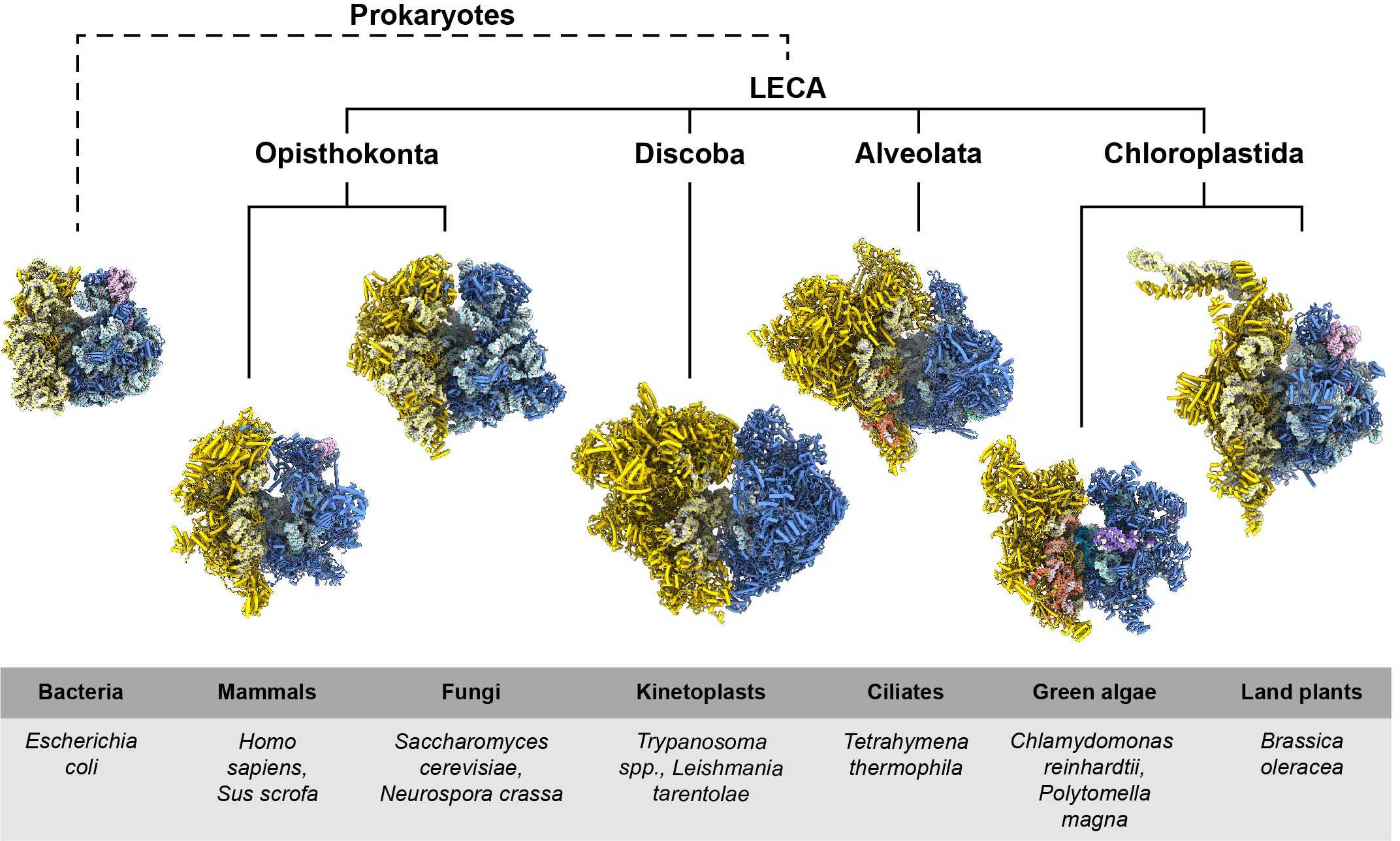
High-resolution structures of the mammalian mitoribosome solved in our group in collaboration with the group of Ruedi Aebersold (ETH Zurich) (external page Greber, Bieri, and Leibundgut et al. 2015), and by the Ramakrishnan Group (external page Amunts and Brown et al. 2015), using cryo-electron microscopy (cryo-EM), provided first insights into the unique features of mitoribosomes. Since then, cryo-EM structures of mitoribosomes from many other organisms have been solved and revealed that mitochondrial ribosomes are not only different from bacterial ribosomes but also feature great diversity amongst themselves (Figure 1). In collaboration with the laboratory of external page André Schneider (University of Bern), we visualised one of the most extremely diverged mitoribosomes, which is found in parasitic protist Trypanosoma brucei (external page Ramrath & Niemann et al. 2018).
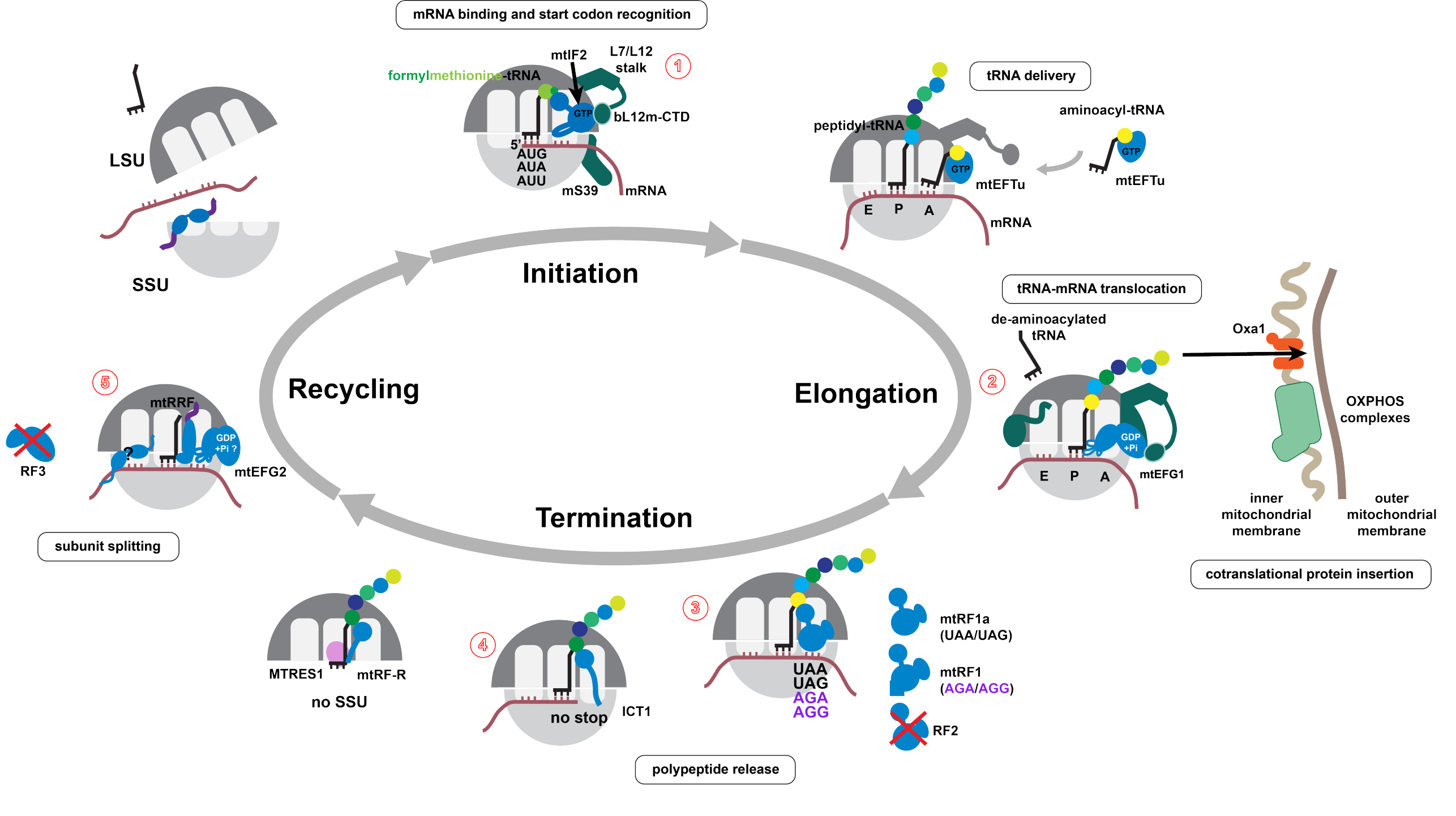
Unique features of the translation cycle in mitochondria
Translation in mitochodria has significantly diverged from the bacterial system. Several key factors, such as initation factor 1 or release factor 2, are absent in mammalian mitochondria. At the same time, new factors, such as mtEFG2 and mtRF1, evolved. In the recent years, many unique features of the mitochondrial translation cycle were uncovered and mechanistically described. Our lab has contributed to this effort with high-resolution cryo-EM structures of several key stages of the translation cycle. Further biochemical analyses and in vivo data optained in cooperation with the research groups of external page Aleksandra Filipovska (University of Western Australia) and external page David Gatfield (University of Lausanne) lead to an increasingly complete picture of translation in mammalian mitochondria (Figure 2).