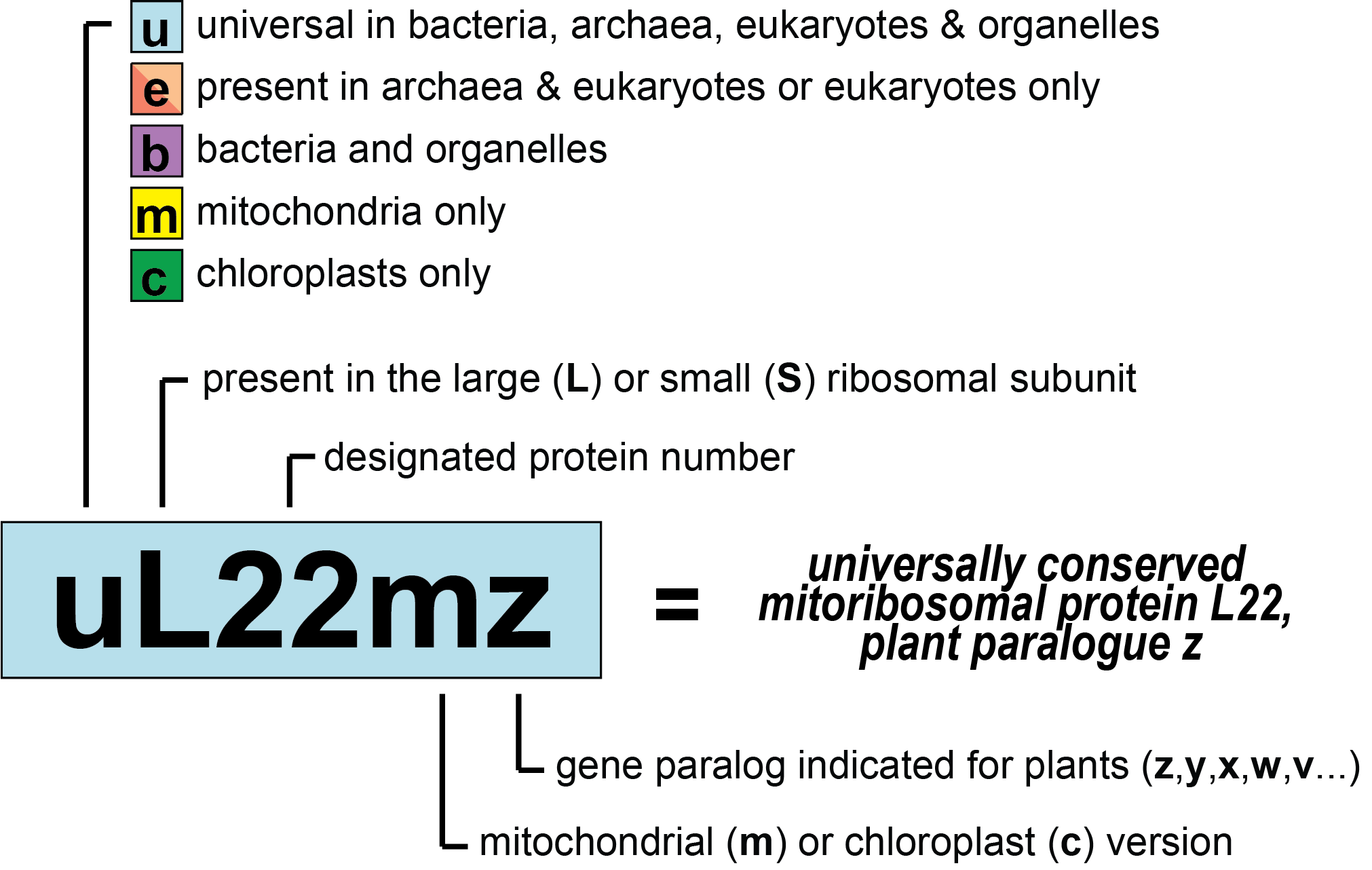
Nomenclature of Ribosomal Proteins
Here we provide information on the updated nomenclature for ribosomal proteins, including helpful tables and PyMOL sessions.
Why do we need a new naming system?
Historically, several unrelated ribosomal proteins from different species were named similarly, while some related proteins were given different names. This had the potential to lead to confusion in the research community. With a multitude of ribosome structures now available at atomic level, it became possible to unambiguously assign most ribosomal proteins to a family based on both sequence and structural homology.
In order to eliminate the naming discrepancies, a consortium of structural biologists and biochemists proposed a new naming system for ribosomal proteins that is universally applicable and also takes into account the historical names wherever possible (external page Ban et al. 2014). The naming system was later extended to include the mitochondrial and chloroplast ribosomal proteins (external page Greber and Ban 2015 and external page Bieri et al. 2016) and recently further expanded to consider the paralogues in plants, which are important for this field (external page Scarpin et al. 2023).
To make the use of the new nomenclature system (Figure 1) easier, we provide an overview figure (Figure 2) as well as detailed conversion tables and PyMOL sessions below.
Consistent with UniProt and the PDB
To consolidate the naming of the > 56,000 ribosomal protein entries in the external page UniProt database, the updated naming system is now implemented by UniProt and linked to the Protein Data Bank (external page PDB).
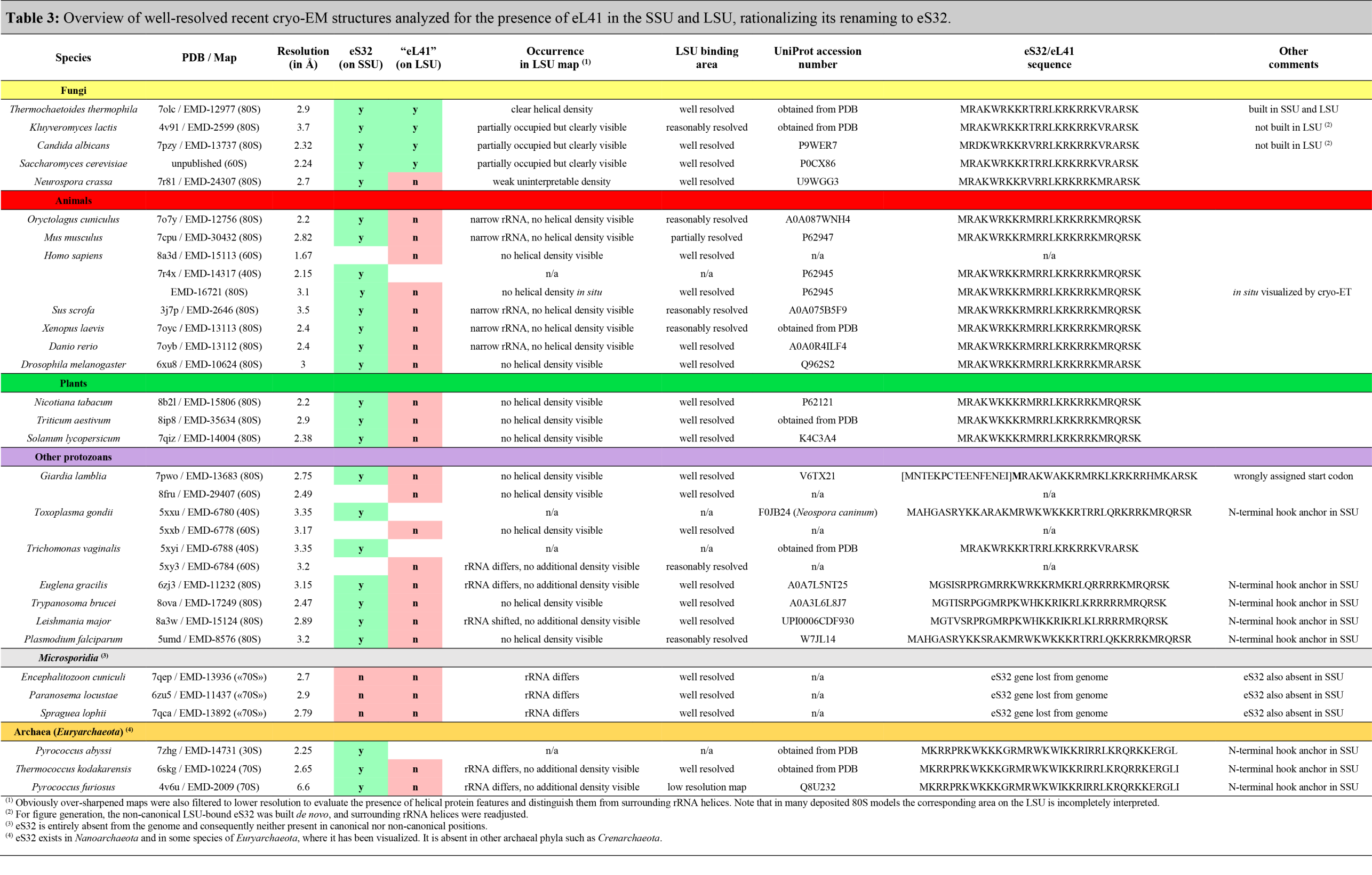
Currently, we suggest only one notable change to the originally proposed naming system: As protein eL41 is always found associated with the small ribosomal subunit (SSU) in archaeal and eukaryotic ribosome structures and is only present in a few fungi at the secondary binding side in the periphery of the large subunit (LSU), we propose to rename eL41 to eS32 to correctly account for its ribosomal location and evolutionary origin (see Table 3, Figure 4 and Figure 5 and accompanying text below).
Updated Nomenclature for Plant Ribosomal Proteins
Since the original proposal of the evolutionary-based naming system, the new names of ribosomal proteins have been widely adopted. However, for plant biologists the naming system turned out to be less useful due to the existence of multiple paralogous copies of ribosomal genes in plants, which require a specific indication of the gene locus. For this reason, external page Lan et al. (2022) and external page Scarpin et al. (2023) proposed a plant-specific extension of the naming system, and the plant community generally agreed to follow the nomenclature suggested by Scarpin et al. (2023). The new convention is also in accordance with public databases such as The Arabidopis Information Resource (external page TAIR), external page MaizeGDB and the external page Plant Cytoplasmic Ribosomal Proteins Database.
UniProt adopted this nomenclature with a minor change: In the UniProt database, no upper-case C or M is given in the suffix to specify organelle-encoded proteins as in Scarpin et al. (2023), because this would be inconsistent with the nomenclature of ribosomal proteins from non-plant sources and is a species-dependent trait. Note that until the new plant ribosomal protein nomenclature is well-established in the field, previous common ribosomal protein names should be additionally indicated in parentheses, and unique gene locus IDs should also be included.
Shortcuts to Tables and PyMOL Sessions
- Table 1a. SSU of bacterial, fungal, and mammalian (mito)ribosomes
- Table 1b. LSU of bacterial, fungal, and mammalian (mito)ribosomes
- Table 2a. SSU of plant cytosolic and organellar ribosomes
- Table 2b. LSU of plant cytosolic and organellar ribosomes
- Table 3. Presence of eS32(eL41) in the SSU/LSU
- How to use the PyMOL sessions
- PyMOL Session: Fungal cytosolic 80S ribosome
- PyMOL Session: Mammalian cytosolic 80S ribosome
- PyMOL Session: Archaeal 70S ribosome
- PyMOL Session: Bacterial 70S ribosome
- PyMOL Session: Chloroplast 70S ribosome
- PyMOL Session: Mammalian 55S mitoribosome
- PyMOL Session: Fungal 74S mitoribosome
- PyMOL Session: Plant cytosolic ribosome
- PyMOL Session: Plant mitoribosome
- PyMOL Session: Algae mitoribosome
Information about the Conversion Tables
- Proteins are colored based on their taxonomic range (cyan: universal, purple: bacterial, orange: archaeal/eukaryotic, red: eukaryotic, yellow: mitochondrial, and green: chloroplast).
- Abbreviations for the occurrence: A: Archaea, B: Bacteria, E: Eukaryotes, m: Mitochondria, c: Chloroplasts.
- Last update: April 2024
Homologues or not?
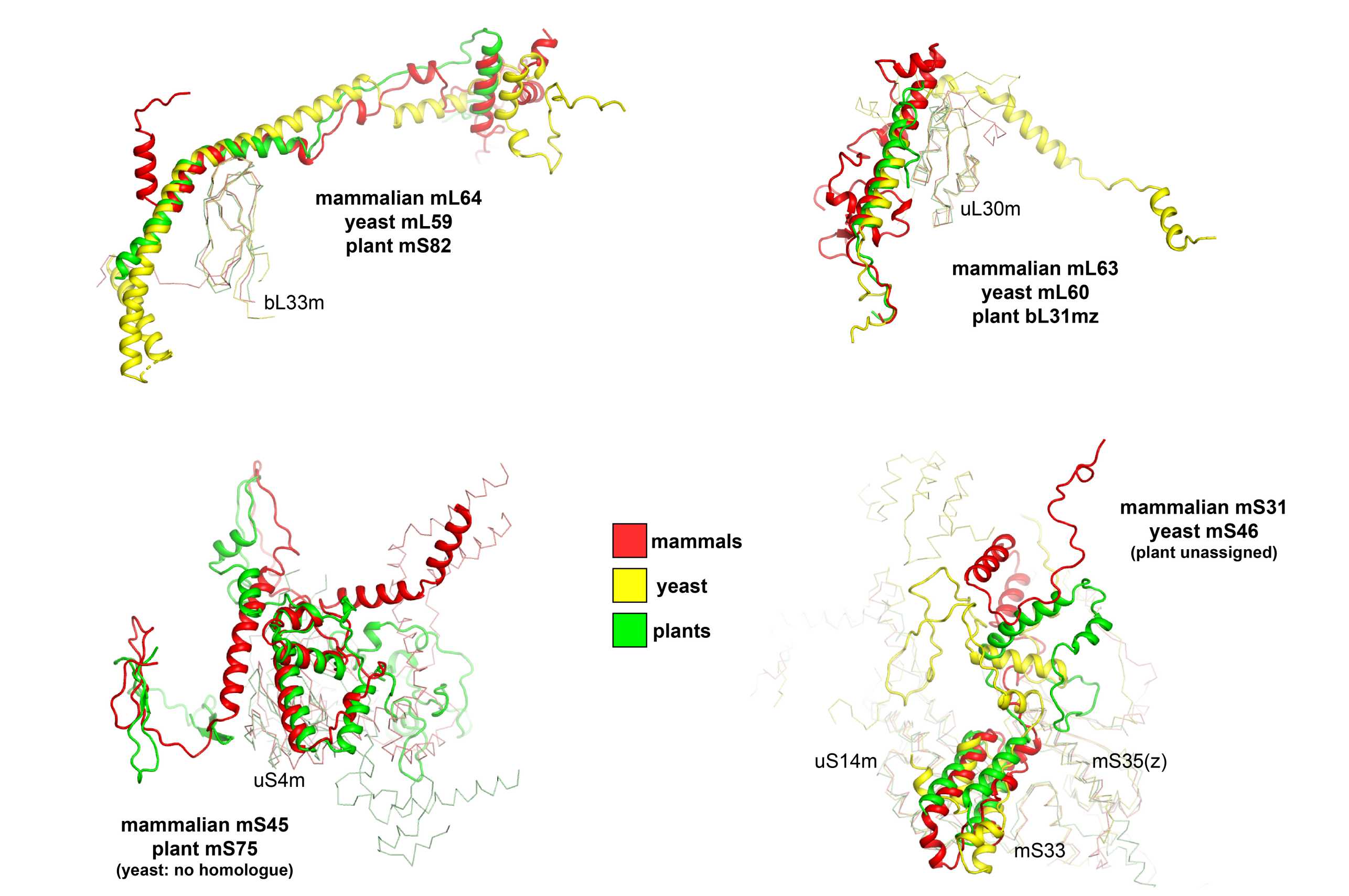
Due to the evolutionary diversity of mitoribosomes and the lack of structural, biochemical and genetic information for many of them, the assignment of novel proteins is still ongoing. Exceptions are mammalian, yeast and plant mitoribosomes, the latter only being available since recently (external page Waltz et al., 2020). While most mammalian, yeast and plant mitoribosome-specific proteins can be unambiguously named based on both structural and sequence homology, for some the evolutionary relationship still remains unclear (Figure 3). For these, although some α helices colocalize in the structures after superposition, no obvious sequence homology can be detected. As UniProt relies on sequence homology to determine the evolutionary relationship of proteins, it was decided to attribute these with different names.
Renaming of eL41 to eS32
Historically, the non-essential ~25 amino acid eL41 peptide (external page Dresios et al. 2003) was named based on its copurification with the LSU of baker’s yeast (Saccharomyces cerevisiae) (external page Otaka & Osawa, 1981 and references therein). However, based on recent structural evidence (Table 3), it became clear that in those eukaryotic and archaeal species where the L41 gene is present in the genome, eL41 is always associated with the SSU at the subunit interface behind the functionally important rRNA helix h44, while its contact with the LSU at this position is only minor (external page Kisly and Tamm 2023), rationalizing it’s renaming to eS32 (Table 3, Figure 4). Furthermore, in archaea and some protists, eS32 carries an N-terminal extension that proceeds further into the SSU rRNA and may serve as an additional hook or anchor (Table 3, Figure 4C).
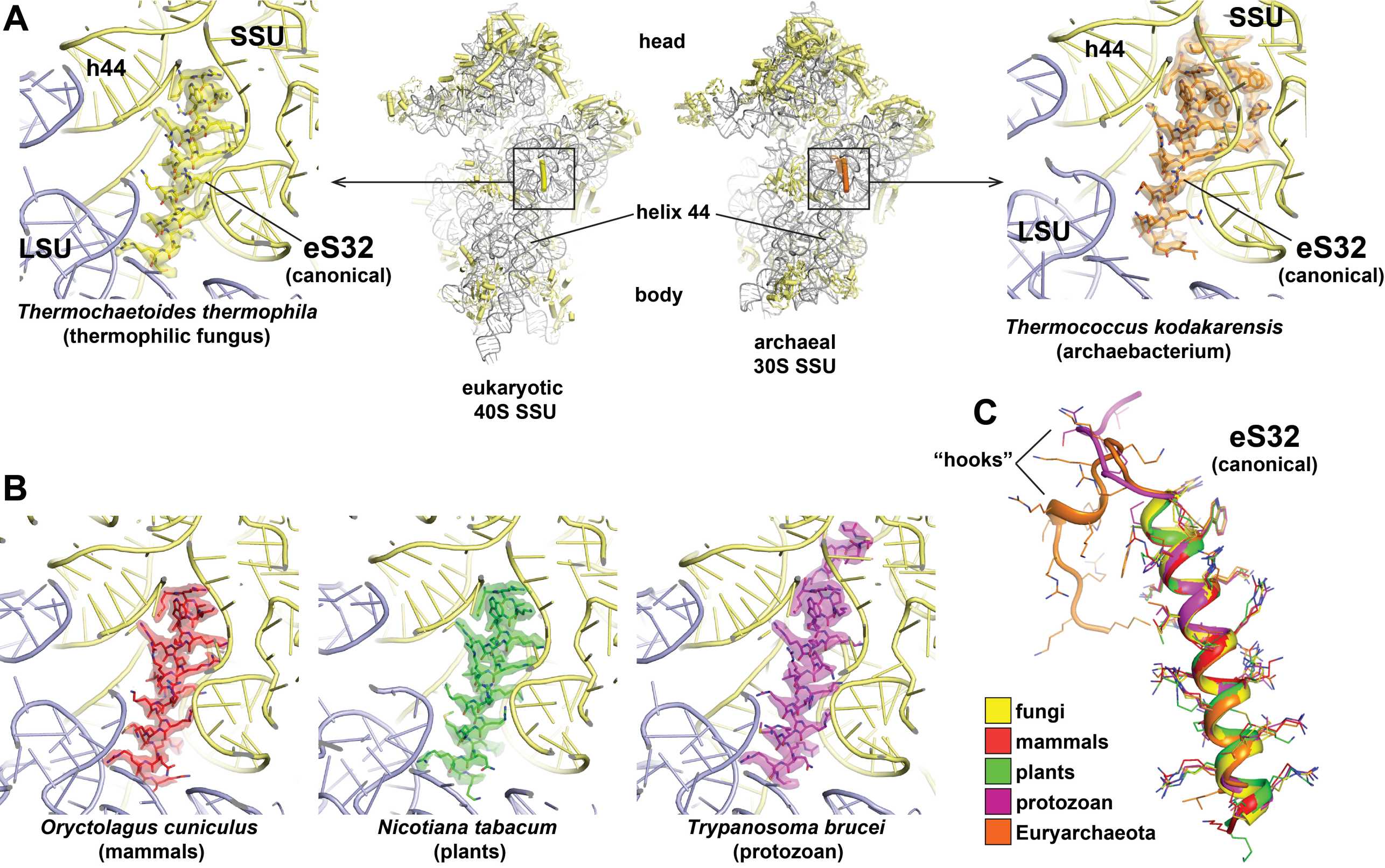
Only in a few fungal species, including baker’s yeast, a second eS32 peptide can be found in some EM maps at a non-canonical rRNA binding pocket at the periphery of the LSU (Kisonaite et al. 2023, Table 3, Figure 5A & 5B). In structurally well-characterized ribosomes of all other eukaryotes, the corresponding rRNA area is occluded or adopts a different shape, and in archaea is even entirely absent (Figure 5C).
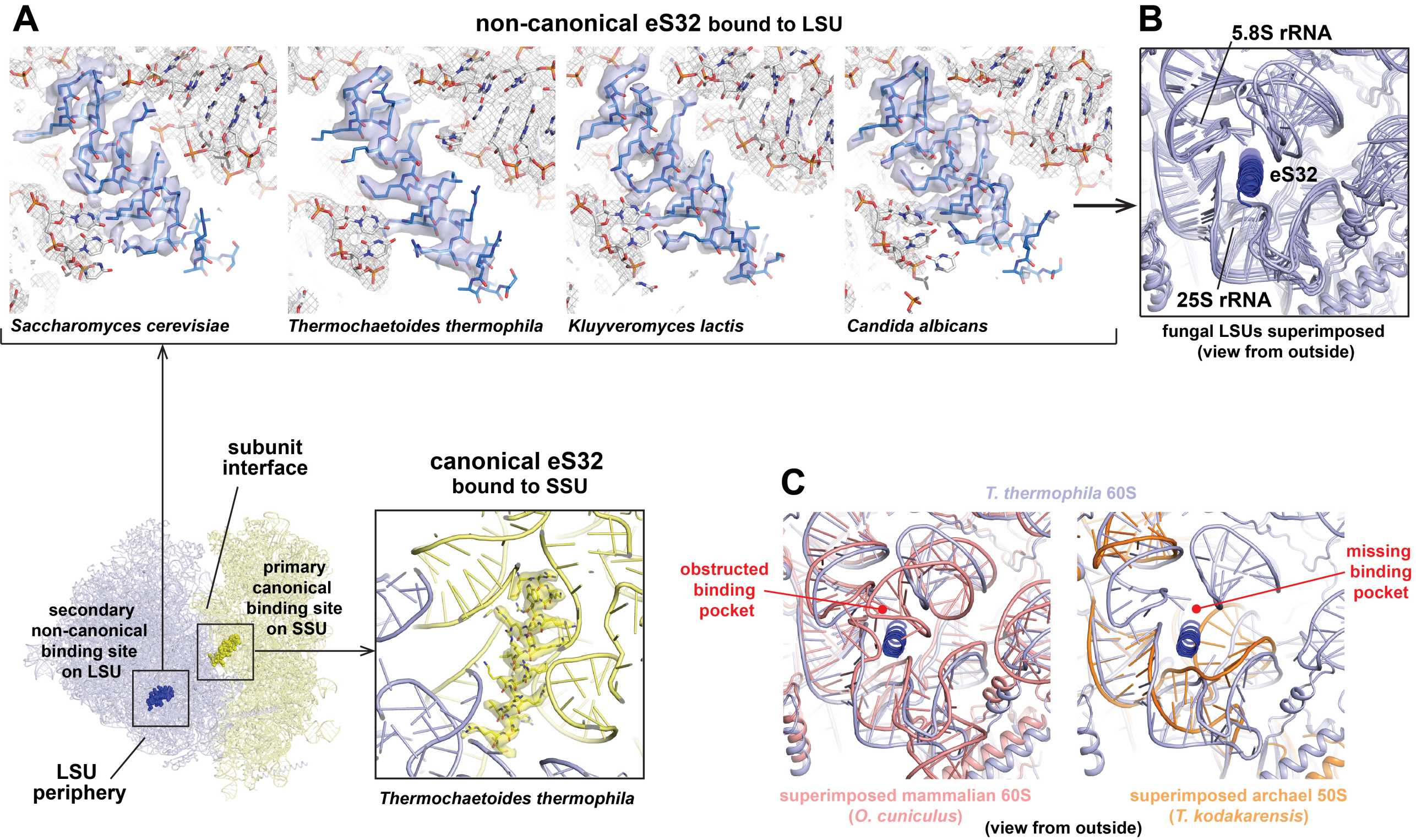
From these observations, it is plausible to assume that eS32 is a SSU protein of archaeal phylogenetic origin, and that its recruitment to the LSU of some fungi was a secondary event. The density for eS32 in EM or X-ray maps is typically also weaker compared to the areas in the vicinity, which indicates a sub-stoichiometric binding and that it could easily dissociate during purification depending on chosen buffer conditions.
In summary, eukaryotic ribosomal proteins were first assigned in ribosomal subunits of baker’s yeast (Otaka & Osaka, 1981 and references therein), which regarding the presence of eS32 in the LSU is rather the exception than the rule. Likely, eS32 was overseen as a component of the SSU for a long time either because it was lost from the subunit during purification or due to its small size and highly basic nature (pI=13), both of which could lead to issues for detection depending on the experimental setup. Only with the resolution revolution in cryo-EM, which allows structure determination of ribosomes from numerous species under more physiological conditions, and recently even in situ (Xing et al. 2023), it became apparent that eL41 is actually a component of the small ribosomal subunit, reasoning its renaming to eS32.
eS32, bS22 or mS38?
Apart from eS32 found at the highly conserved canonical binding site at the cytosolic SSU, small helical proteins located behind helix h44 of the SSU were also detected in mammalian and fungal mitoribosomes (named mS38, Table 1a) and in Actinobacteria and Bacteroidetes (named bS22 in external page Hentschel et al. 2017, external page Jha et al. 2021, and external page Sharma et al. 2023). However, although the binding to the rRNA is also mediated by numerous positively charged residues and on the sequence level mS38 and bS22 are distantly related, they do not share obvious homology with eS32 and are positioned differently in the SSU (Hentschel et al. 2017). Therefore, a common evolutionary origin of these helical peptide proteins remains unclear and their names are kept separate.
PyMOL Scripts and Sessions
We provide a number of scripts and sessions for "external page The PyMOL Molecular Graphics System" for ribosomes from different organisms. The updated nomenclature is implemented through the use of both the old and new ribosomal protein names.
The X-ray structure of the yeast cytosolic ribosome (PDB ID external page 4V88, large subunit B in the asymmetric unit, external page Ben-Shem et al. 2011) served as template to which all other structures were aligned in order to allow direct comparison of the proteins from different species.
How do I use the PyMOL scripts?
- Download and install a current version of external page PyMOL (2.2.5 or higher).
- Download the PyMOL session as a .zip file and unzip it
- Double-click the file or open it from within PyMOL (File>Open...)
- Alternatively, you can use the provided script files by copying the text into the command line of the PyMOL GUI. Please find further instructions in the header of each script.
3.0Å crystal structure of the fungal cytosolic 80S ribosome (Saccharomyces cerevisiae)
Download Download PyMOL session file (ZIP, 14.3 MB)
Download Download PyMOL script file (TXT, 10 KB)
PDB ID: 4v88 (molecule B)
external page Ben-Shem, A. et al. Science 334: 1524-1529 (2011)
2.2Å cryo-EM structure of the mammalian cytosolic 80S ribosome (Oryctolagus cuniculus)
Download Download PyMOL session file (ZIP, 15.1 MB)
Download Download PyMOL script file (TXT, 11 KB)
PDB ID: 7o7y
external page Bhatt, P.R., Scaiola, A. et al. Science 372: 1306-1313 (2021)
2.65Å cryo-EM structure of the archaeal 70S ribosome (Thermococcus kodakarensis)
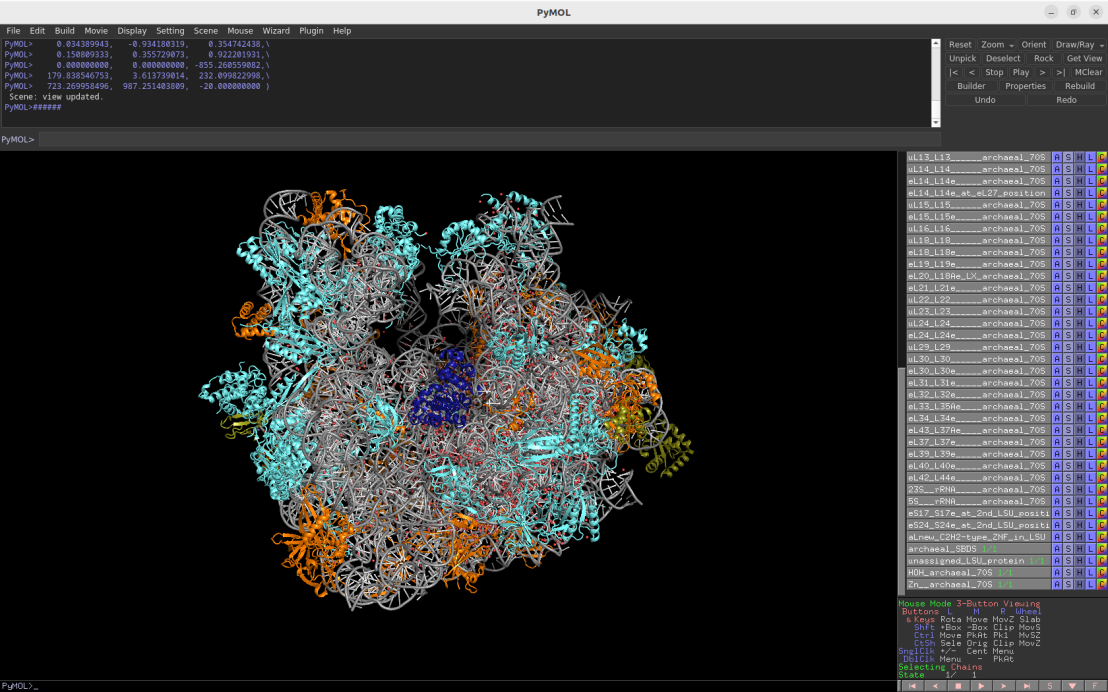
Download Download PyMOL session file (ZIP, 10.3 MB)
Download Download PyMOL script file (TXT, 9 KB)
PDB ID: 6skg
external page Sas-Chen, A. et al. Nature 583: 638-643 (2020)
2.1Å crystal structure of the bacterial 70S ribosome (Escherichia coli)
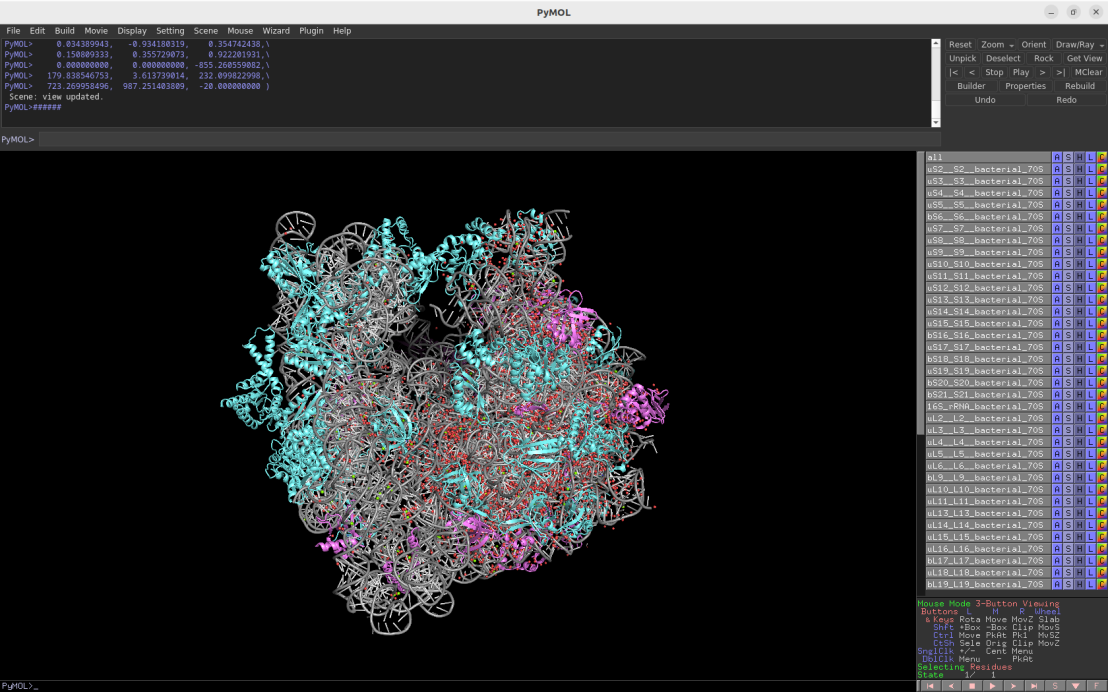
Download Download PyMOL session file (ZIP, 11.2 MB)
Download Download PyMOL script file (TXT, 9 KB)
PDB ID: 4ybb
external page Noeske, J. et al. Nat Struct Mol Biol 22: 336-341 (2015)
3.4Å cryo-EM structure of the chloroplast 70S ribosome (Spinacia oleracea)
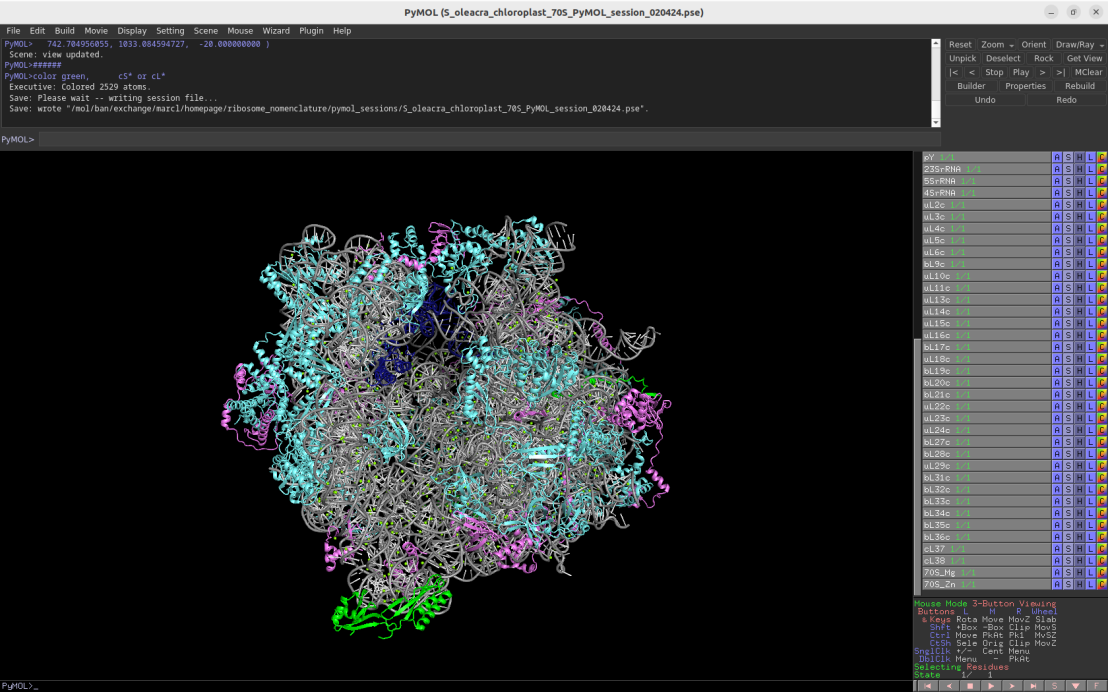
Download Download PyMOL session file (ZIP, 9.5 MB)
Download Download PyMOL script file (TXT, 9 KB)
PDB ID: 5mmm
external page Bieri, P. et al. EMBO J 36: 475-486 (2017)
3.2Å cryo-EM structure of the 55S mammalian mitochondrial ribosome (Sus scrofa)
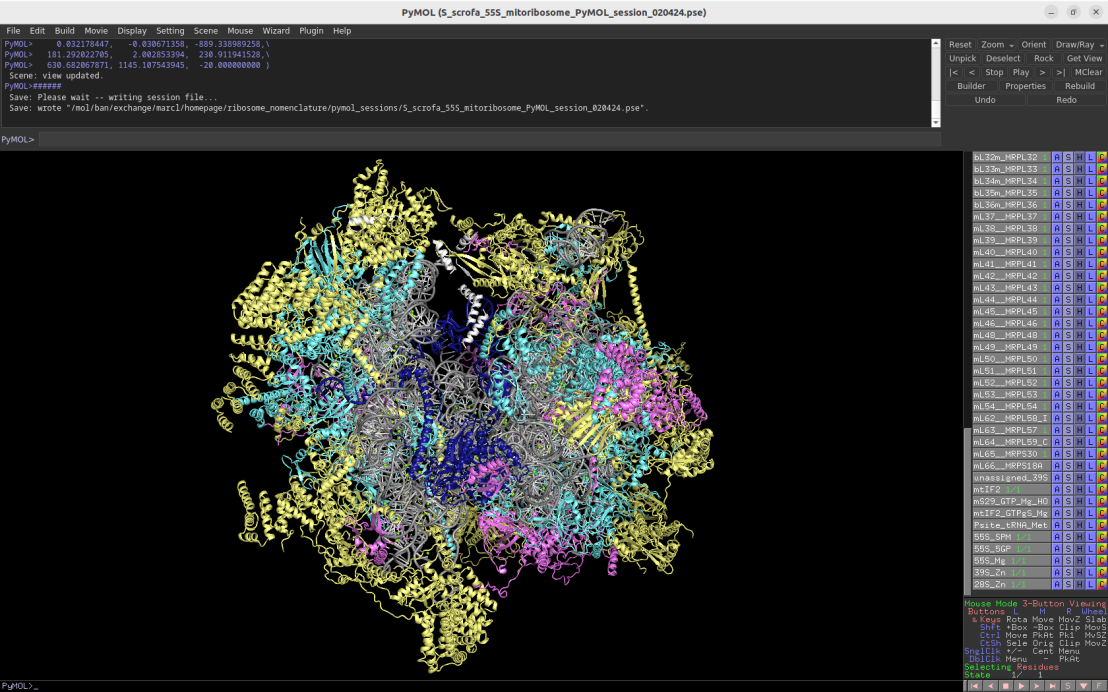
Download Download PyMOL session file (ZIP, 12.1 MB)
Download Download PyMOL script file (TXT, 14 KB)
PDB ID: 6gaw
external page Kummer, E. et al. Nature 560: 263-267 (2018)
3.25Å cryo-EM structure of the fungal 74S mitochondrial ribosome (Saccharomyces cerevisiae)
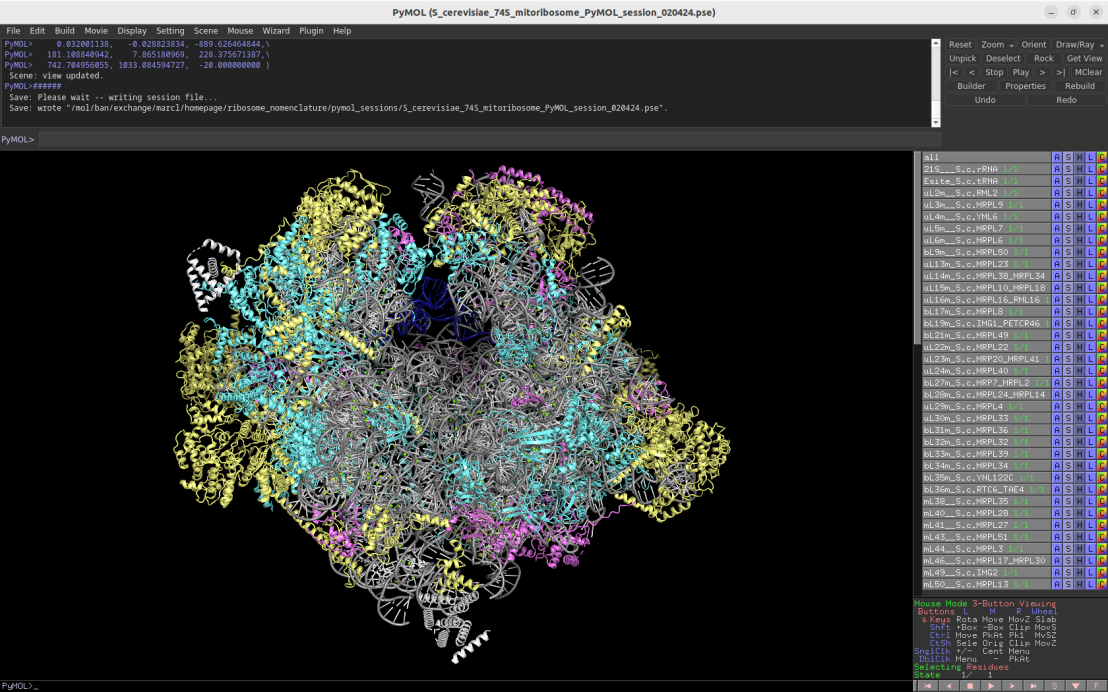
Download Download PyMOL session file (ZIP, 12.7 MB)
Download Download PyMOL script file (TXT, 12 KB)
PDB ID: 5mrc
external page Desai, N. et al. Science 355: 528-531 (2017)
2.2Å cryo-EM structure of the plant cytosolic 80S ribosome (Nicotiana tabacum)
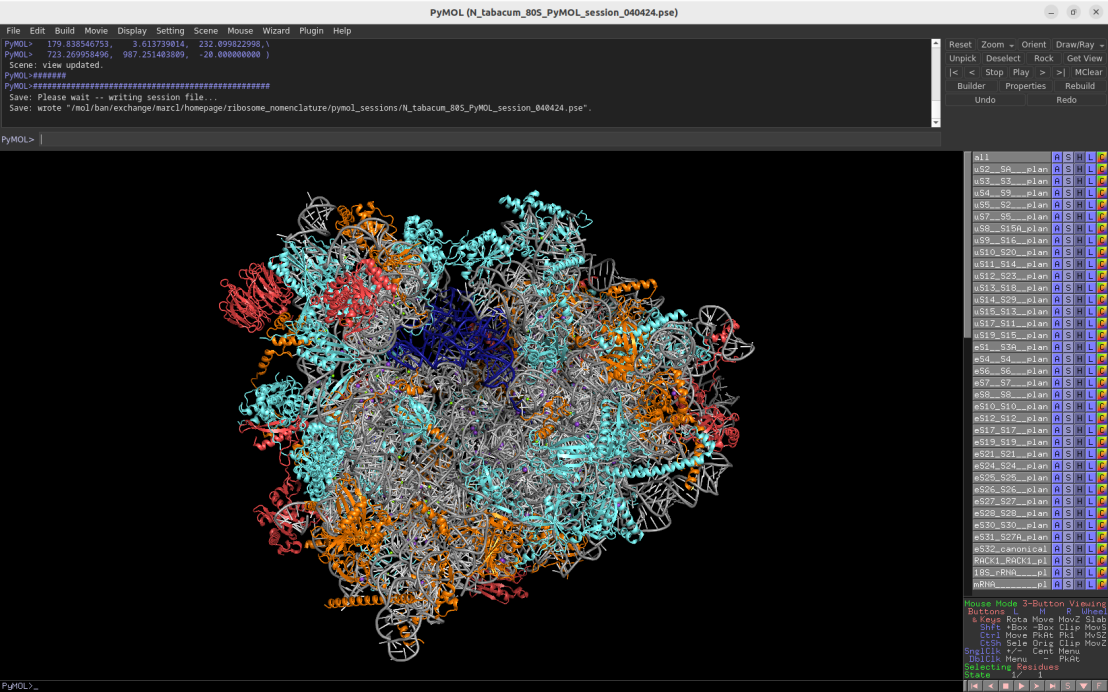
Download Download PyMOL session file (ZIP, 22.4 MB)
Download Download PyMOL script fil (TXT, 11.2 kB) (TXT, 11 KB)
PDB ID: 8b2l
external page Smirnova, J. et al. Nat Plants 9: 987-1000 (2023)
3.86Å cryo-EM structure of the plant mitoribosome (Arabidopsis thaliana / Brassica oleracea)
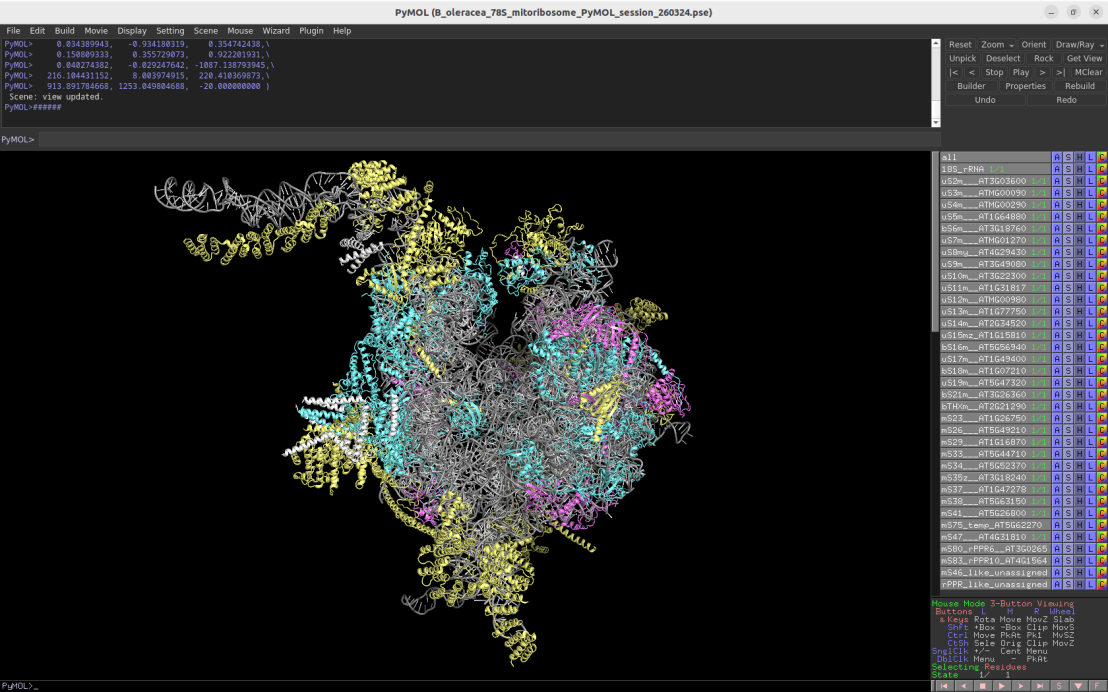
Download Download PyMOL session file (ZIP, 12.2 MB)
Download Download PyMOL script file (TXT, 11 KB)
PDB ID: 6xyw
external page Waltz, F. et al. Nat Plants 6: 377-383 (2020)
2.91Å cryo-EM structure of the algae mitoribosome (Polytomella magna)

Download Download PyMOL session file (ZIP, 12.3 MB)
Download Download PyMOL script file (TXT, 12 KB)
PDB ID: 8a22
external page Tobiasson, V. et al. Nat Commun 13: 6132-6132 (2022)
Help us to improve this page
For questions, error reports and comments please contact .